Python and Linux: Difference between pages
Marc Saric (talk | contribs) |
|||
| Line 1: | Line 1: | ||
= | = My Linux-experience = | ||
I have been using Linux (mostly in a dual-boot environment) since around 1996 or 1997 (Cheapbytes CD from Lehmanns bookstore with Debian 1.24 on a Pentium 133 MHz). | |||
The most recent version of this page has been created with Linux tools alone and is hosted on a Linux box. Some images and older sediments of my site may incorporate Windows-generated images, but nowadays I have switched to Open Source tools to create everything I want to show here. | |||
I am developing Cloud-based software fro a living since my Postdoc days on [http://www.linux.org/ Linux]- and previously [http://www.apple.com/ Apple]-machines, using [[Python]], [https://www.terraform.io/ Terraform], [https://ansible.com/ Ansible], [https://www.docker.com/ Docker], [https://developer.mozilla.org/en-US/docs/Web/JavaScript Javascript], [http://www.mozilla.org Mozilla], [http://www.php.net PHP], [http://www.perl.com Perl],, [http://www.postgresql.org/ PostgreSQL], [http://www.mysql.com MySQL] and various IDEs. | |||
Beside that I was heavily depending on a stable machine to do my [[PhD|Ph.D.]], which was one of the two main reasons for switching from Windows NT to [http://www.novell.com/linux/suse/ SuSE]-Linux and finally [http://fedora.redhat.com/ Fedora] and [https://www.ubuntu.com/ Ubuntu]. | |||
= Pictures = | |||
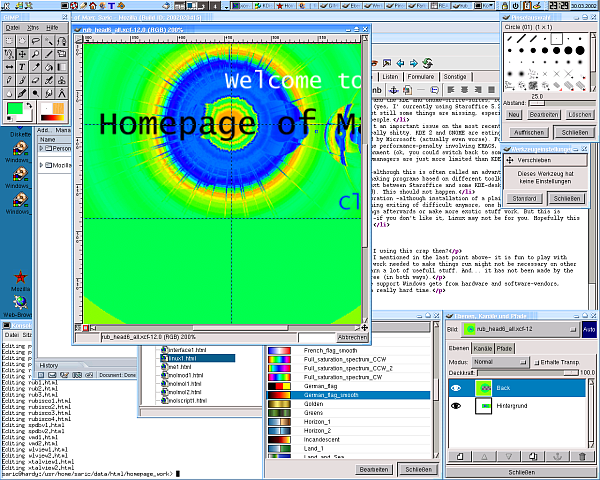
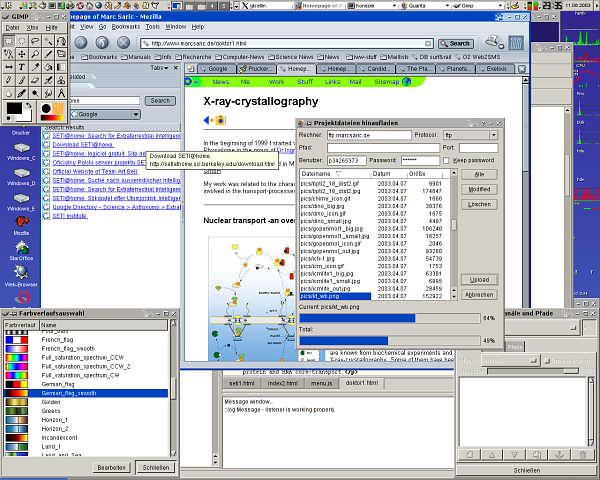
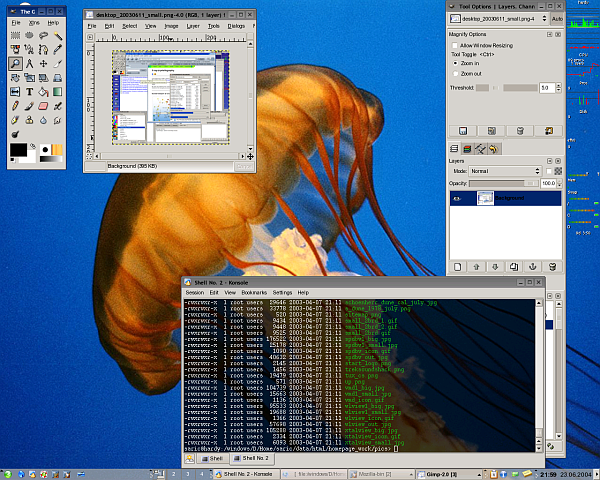
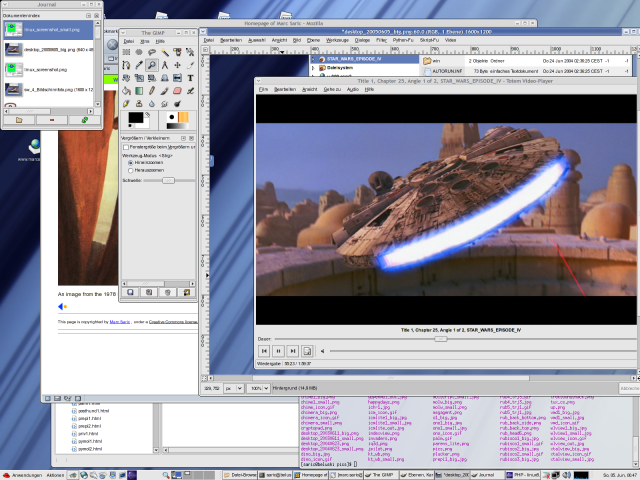
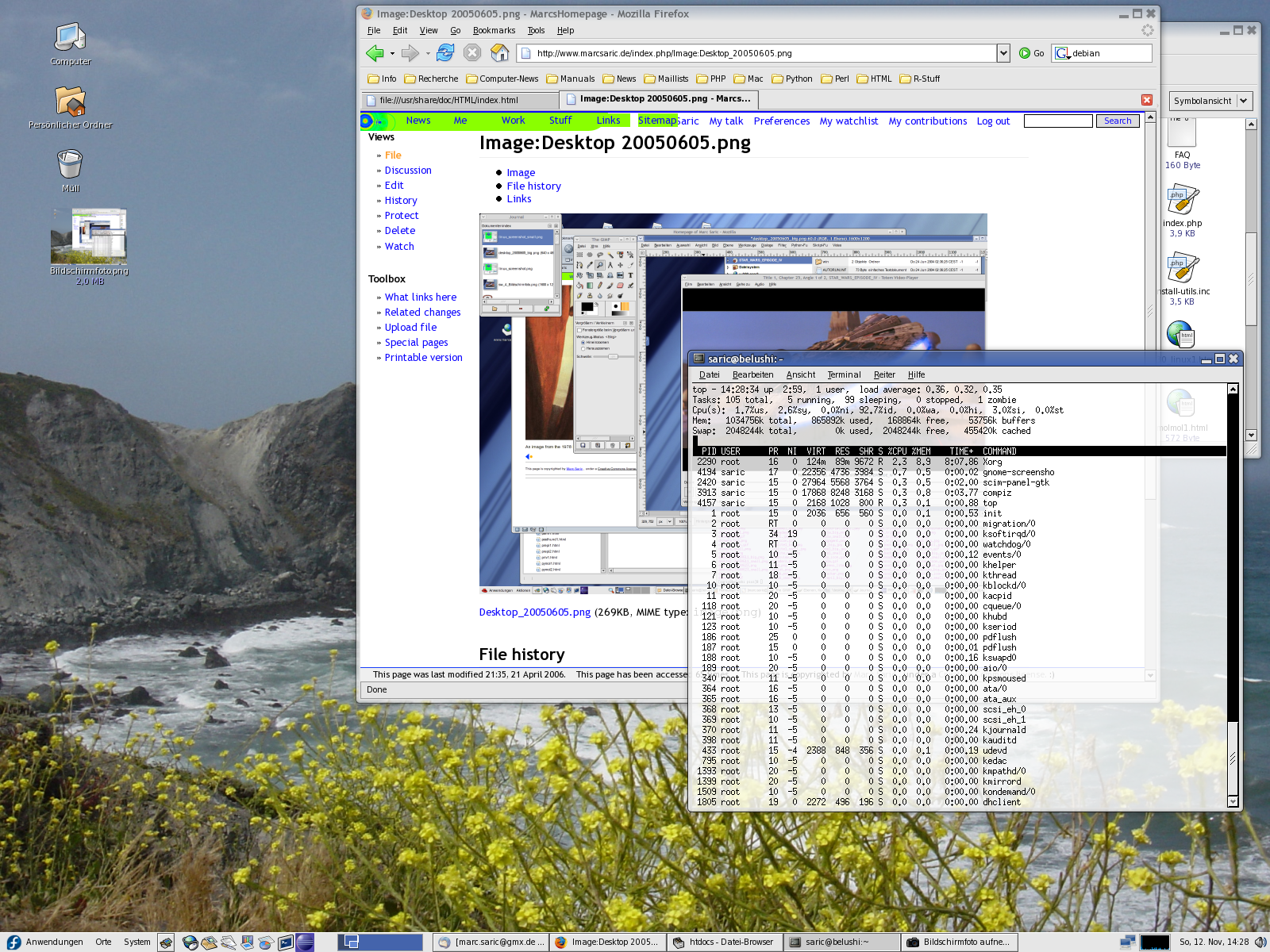
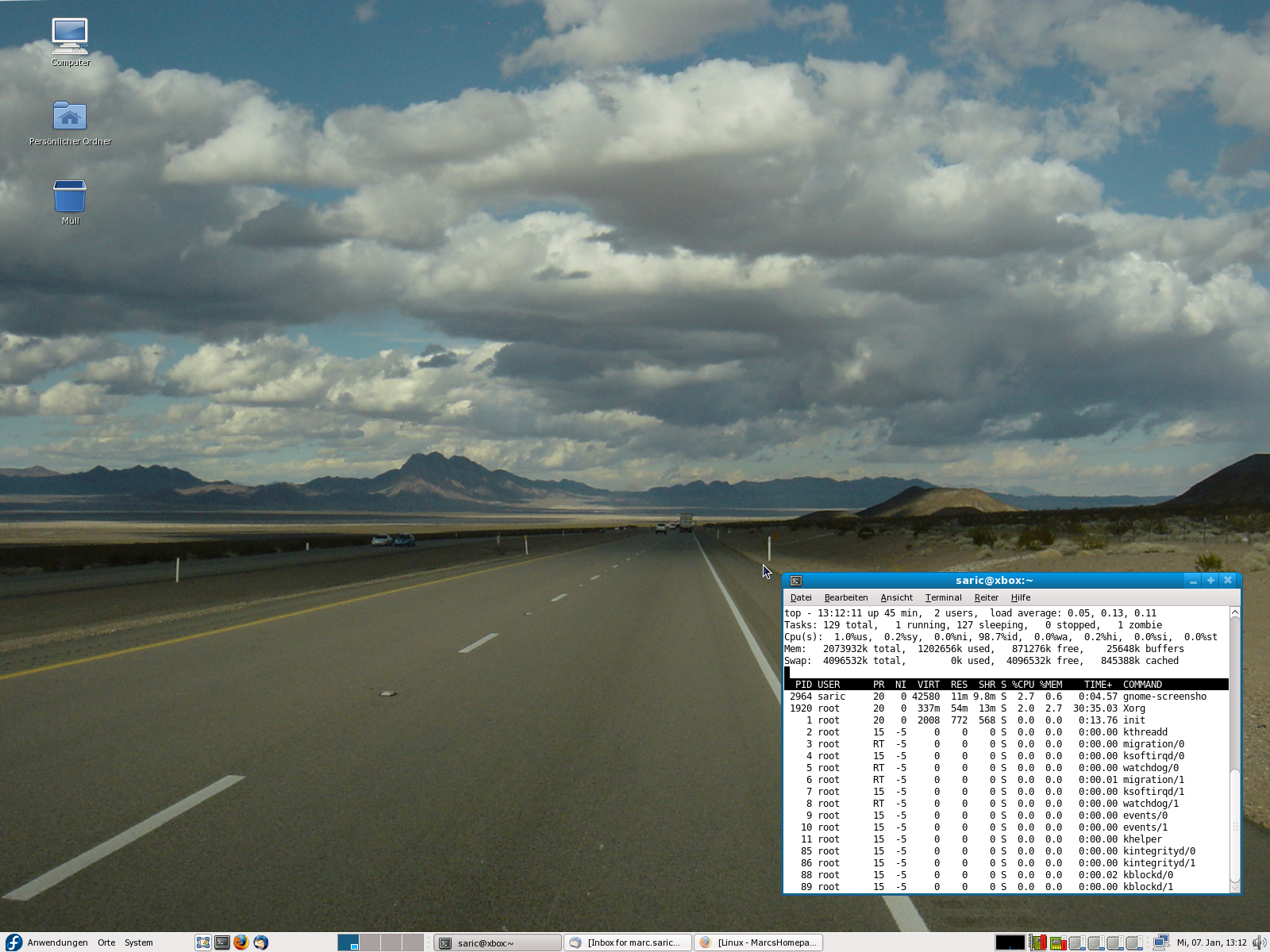
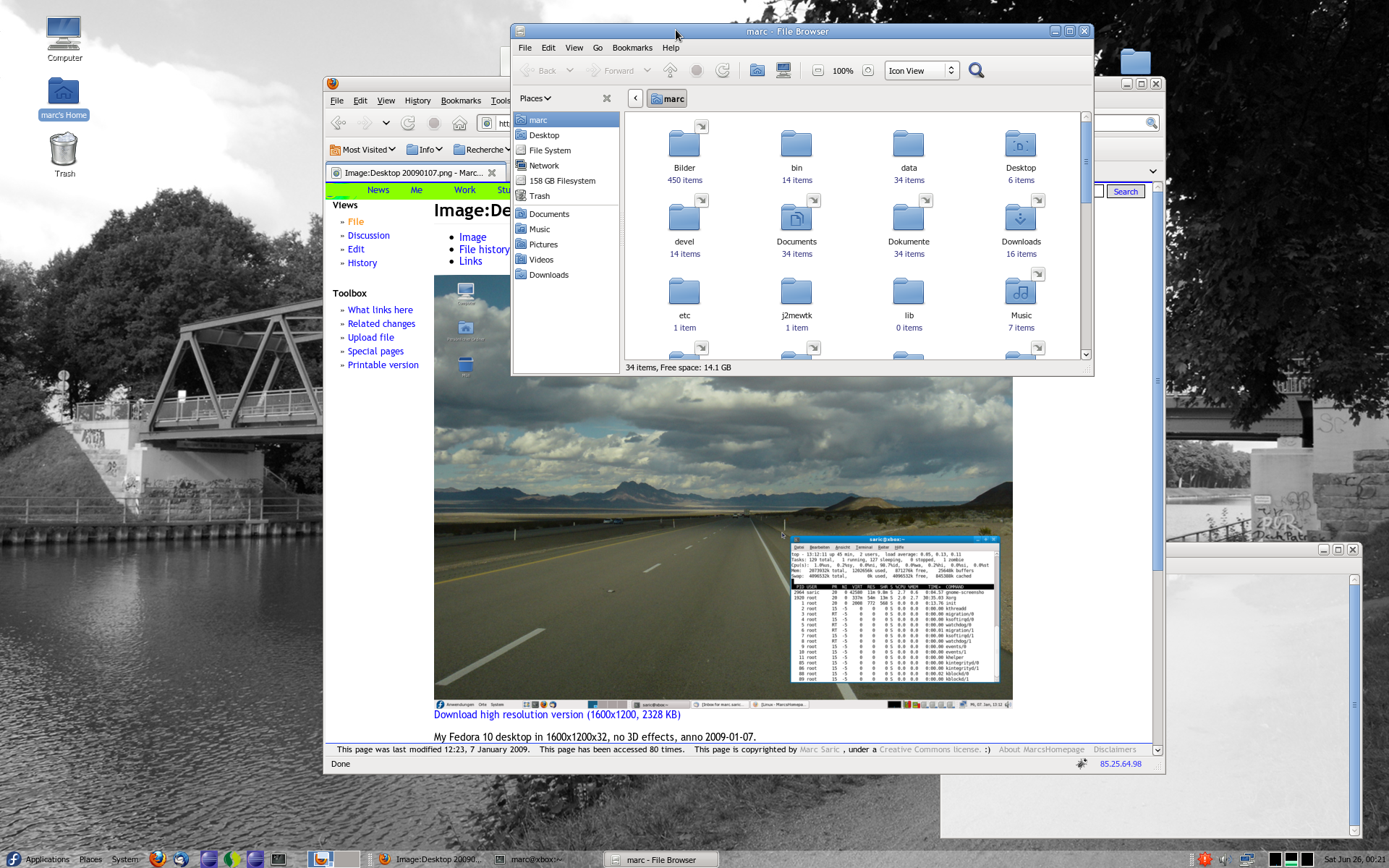
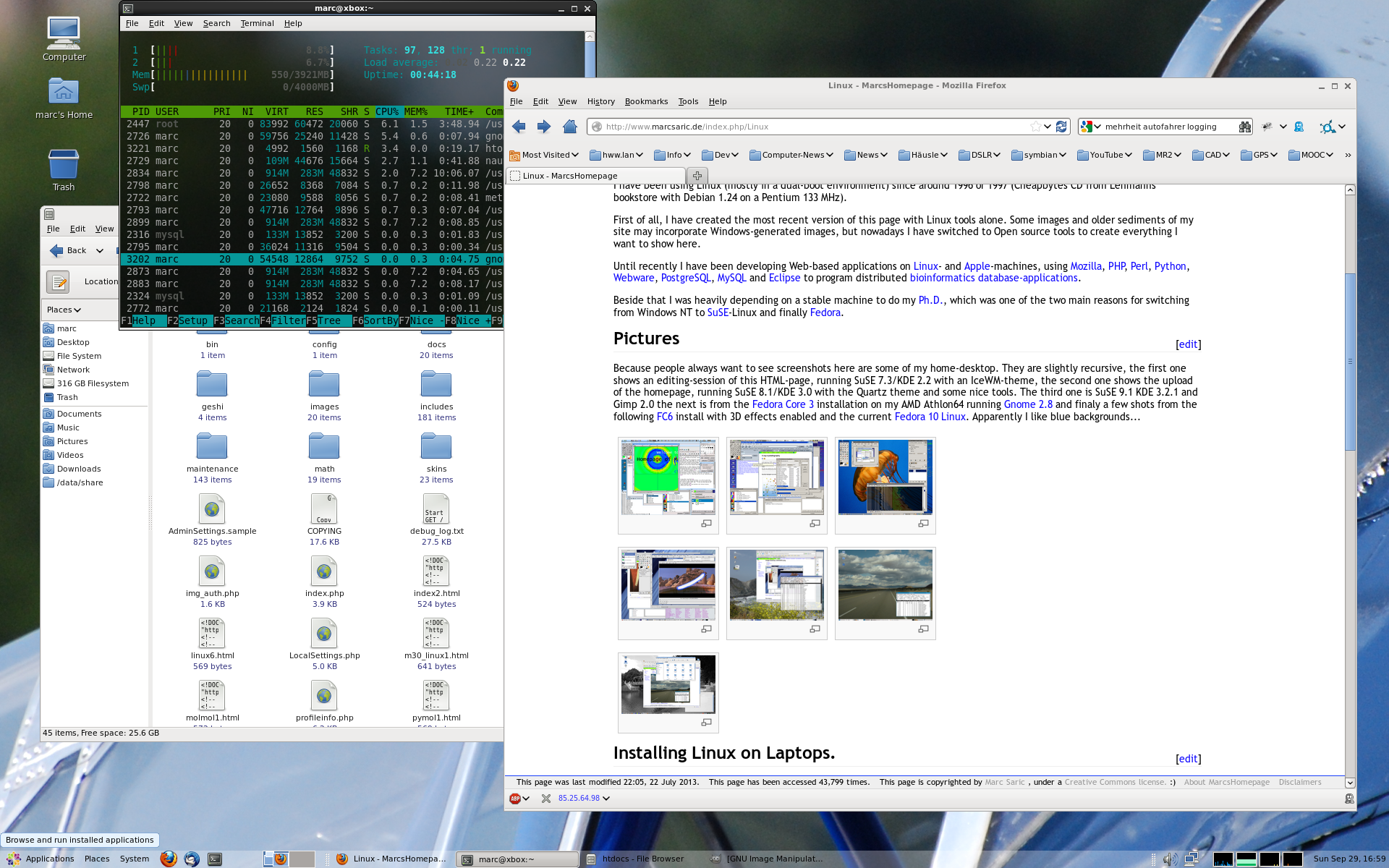
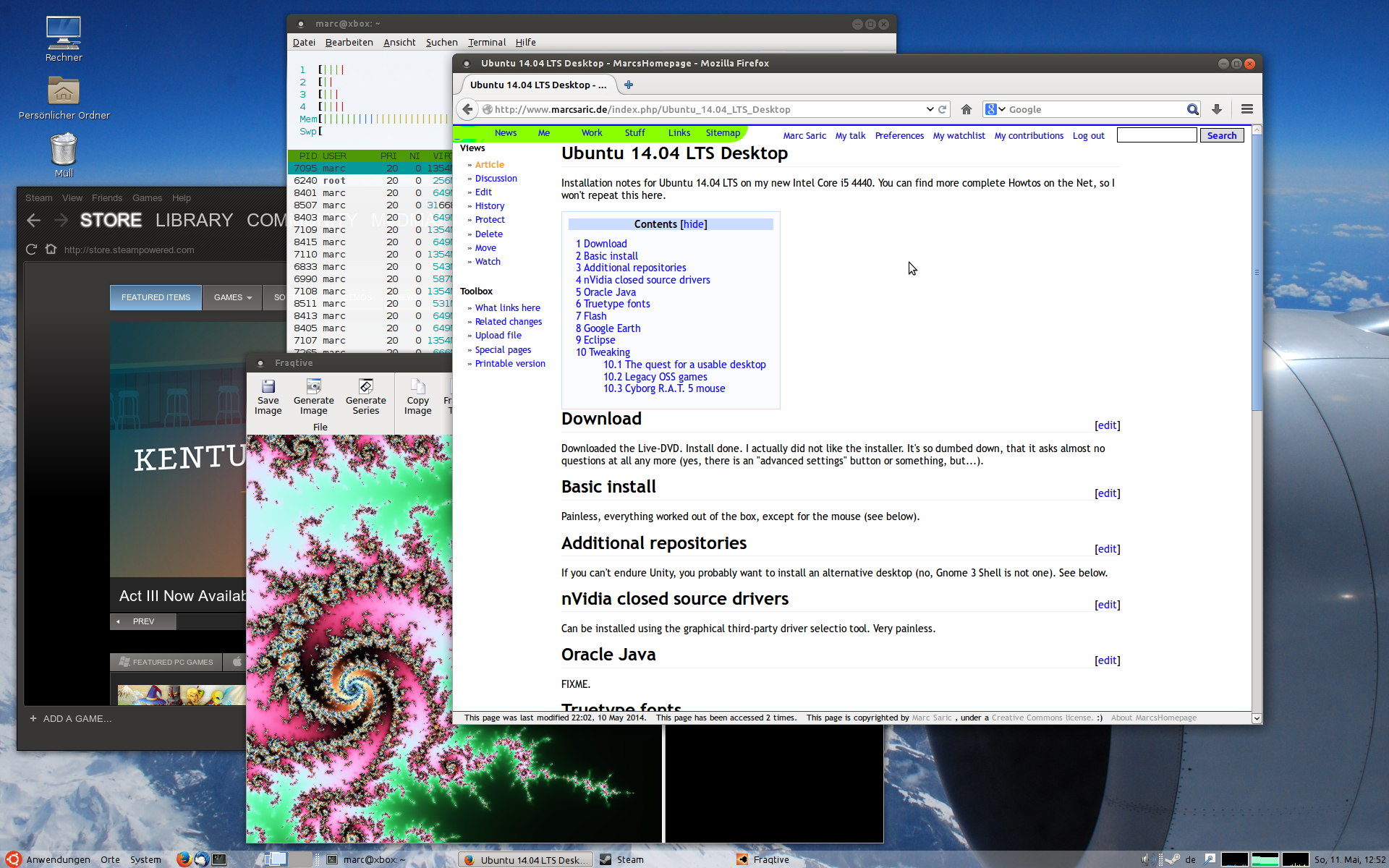
Because people always want to see screenshots here are some of my home-desktop. They are slightly recursive, the first one shows an editing-session of this HTML-page, running SuSE 7.3/KDE 2.2 with an IceWM-theme, the second one shows the upload of the homepage, running SuSE 8.1/KDE 3.0 with the Quartz theme and some nice tools. The third one is SuSE 9.1 KDE 3.2.1 and Gimp 2.0 the next is from the [[Fedora Core 3 Linux|Fedora Core 3]] installation on my AMD Athlon64 running [http://www.gnome.org Gnome 2.8] followed by a few shots from the [[Fedora Core 6 Linux|FC6]] install with 3D effects enabled. Next [[Fedora 10 Linux]] and [[Fedora 13 Linux]] and [[CentOS 6.2|CentOS 6]] and currently [[Ubuntu 14.04 LTS Desktop]]. Apparently I like blue backgrounds... | |||
{| | |||
|[[Image:linux_screenshot.png|thumb|none|230px]] | |||
|[[Image:desktop_20030611.png|thumb|none|230px]] | |||
|[[Image:desktop_20040623_small.png|thumb|none|230px]] | |||
|- | |||
|[[Image:desktop_20050605.png|thumb|none|230px]] | |||
|[[Image:desktop_20061112.png|thumb|none|230px]] | |||
|[[Image:desktop_20090107.png|thumb|none|230px]] | |||
|- | |||
|[[Image:desktop_20100626.png|thumb|none|230px]] | |||
|[[Image:desktop_20130929.png|thumb|none|230px]] | |||
|[[Image:desktop_20140511.png|thumb|none|230px]] | |||
|} | |||
< | = Raspberry Pi = | ||
I got a Raspberry Pi 4 with 2 GB RAM as a gift. Of course, I have to find some useful project for this little machine. | |||
Read my [[Raspberry Pi project]] page for this. | |||
= Installing Linux on Laptops = | |||
Recently, I bought a used Lenovo Thinkpad x230. | |||
Read the review. | |||
* [[Ubuntu 16.04 LTS on Lenovo Thinkpad x230]] | |||
During my work for [http://www.exelixis.com Exelixis], I had an IBM R32 laptop running SuSE 8.1 and during my [[Postdoc|stay]] back at the [http://www.uni-tuebingen.de university], I had a [http://cdgenp01.csd.toshiba.com/content/product/pdf_files/detailed_specs/satellite_M30_Series.pdf Toshiba M30-344<small>[pdf-file]]</small>, which I used with [http://www.novell.com/linux/suse/ SuSE] 9.0 and 9.1. | |||
Short note: Both pages will not get updates, as I don't own those laptops any more. | |||
Blue pill or red pill... | |||
* [[SuSE 8.1 on an IBM R32]] | |||
* [[SuSE 9.0 on a Toshiba M30-344]] | |||
= Gentoo Linux on Small Systems = | |||
Installing [http://www.suse.de one] [http://www.mandrakesoft.com/ of the] [http://www.debian.org more recent] [http://www.redhat.com distros] on an old system (aka [[User:MarcSaric/Private|my 300 MHz PII]]) is not really fun. | |||
Except for the fact, that a small hard disk is quickly filled up by a load of gunk you might not need at all (but which is installed anyway, if you don't select rpm-packages by hand), those distros tend to be painfully slow (the current version of Quanta IS dead-slow even on my AMD Duron 700 (yess, CPUs have arrived ad [http://www.xtremesystems.org/forums/showthread.php?t=42655 6.0 GHz meanwhile], I know)), and take eons to load the desktop (why? I was running Linux on my 486 ages ago). | |||
There seems to be no real hope for a fast-and-small version of SuSE, therefore I had to look for something else. | |||
'''Enter Gentoo-Linux:''' (I have to admit, that almost everything which follows, has been stolen from an email I got from a [http://www.professa.de friend], who setup all this on his machine ;-)). | |||
Anyway, if you are interested in my installation-notes for a small-footprint Gentoo-system, [[Gentoo Linux|read on]]. | |||
= Fedora = | |||
Several years ago I switched from my long-time favorite [http://www.novell.com/linux/ Linux-distribution] to [http://fedora.redhat.com Fedora Core 3]. | |||
== Installation notes for [[Fedora Core 3 Linux]] == | |||
Permian period. My first stab at Fedora Linux after I switched from SuSE. Quite some manual effort here compared to more recent distros. | |||
== Installation notes for [[Fedora Core 6 Linux]] == | |||
Jurrassic age. One of the more unstable Fedora releases. | |||
== Installation notes for [[Fedora 8 Linux]] == | |||
Stone age. A better one (compared to the previous install). | |||
== Installation notes for [[Fedora 10 Linux]] == | |||
Very outdated. Some major havoc with Sun Java. Apart from that, the install becomes more and more <strike>boring</strike> mainstream | |||
== Installation notes for [[Fedora 13 Linux]] == | |||
Outdated. New install, new glitches, among them one, which cropped up in the current Ubuntu install. | |||
== Installation notes for [[Fedora 14 Linux]] == | |||
The last Fedora. I switched to CentOS afterwards. | |||
= CentOS = | |||
== Installation notes for [[CentOS 4.3]] == | |||
Some time ago I had to setup a 64-bit 4-way server and decided to give CentOS a shot. Having ordered a floppy-drive ("who really needs that nowadays") immediately paid off, because the RAID-Controller was not recognized and a newer kernel-module had to be installed from a boot-floppy (not a problem, unless you can't use one). | |||
== Installation notes for [[CentOS 6.2]] == | |||
After everyone decided, that the new GUI for Linux should look like a Smartphone, I moved from Fedora to CentOS 6 (which also has a longer life-time and does not force you to install a new OS every 1,5 years or so). | |||
= Ubuntu Linux = | |||
== Installing Ubuntu 7.10 == | |||
Read my review on installing [[Ubuntu 7.10 Gutsy Gibbon]]. I did this recently for one of my older machines, because the old Ubuntu-version was no longer supported. | |||
== Installing Ubuntu 12.04 LTS 64 bit for XBMC == | |||
I have built a [http://xbmc.org/ XBMC] based [https://en.wikipedia.org/wiki/Home_theater_PC Home Theater PC (HTPC)] using [http://releases.ubuntu.com/precise/ Ubuntu 12.04 LTS 64 bit] on an Intel platform. | |||
Read my [[XBMC HTPC]] installation notes. | |||
== Installing Ubuntu 14.04 LTS 64 bit == | |||
My last PC died from [https://en.wikipedia.org/wiki/Capacitor_plague Capacitor plague] after more than 6 years, so I had to buy a new one. CentOS 7 was not quite ready, so I choose the new Ubuntu LTS. Read my [[Ubuntu 14.04 LTS Desktop]] installation notes. | |||
== Upgrading to Ubuntu 16.04 LTS 64 bit == | |||
As the update client kept nagging me, that a new version is out, I followed that advice and updated the existing system to Ubuntu 16.04 LTS. Read my [[Ubuntu 16.04 LTS Desktop]] upgrade notes. | |||
== Upgrading to Ubuntu 18.04 LTS 64 bit == | |||
I did an upgrade to Ubuntu 18.04 LTS. Reading my [[Ubuntu 18.04 LTS Desktop]] page is worthless, as it is empty (nothing to fiddle around with there). | |||
== Upgrading to Ubuntu 20.04 LTS 64 bit == | |||
I did an upgrade to Ubuntu 20.04 LTS. There is nothing to read there [[Ubuntu 20.04 LTS Desktop]]. | |||
= Gaming = | |||
{| | |||
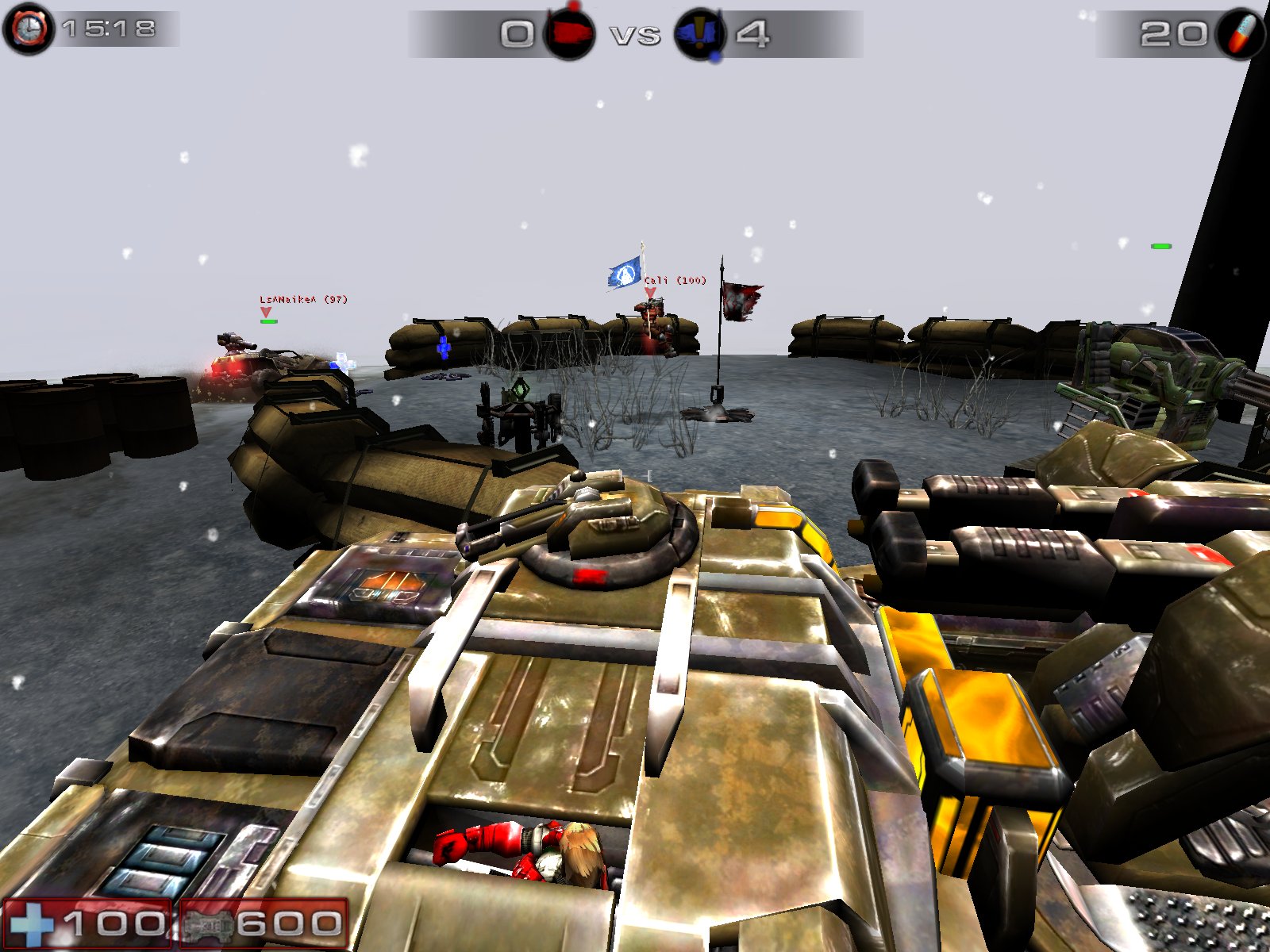
|[[Image:ut2004.jpg|thumb|UT2004 VCTF|150px]] | |||
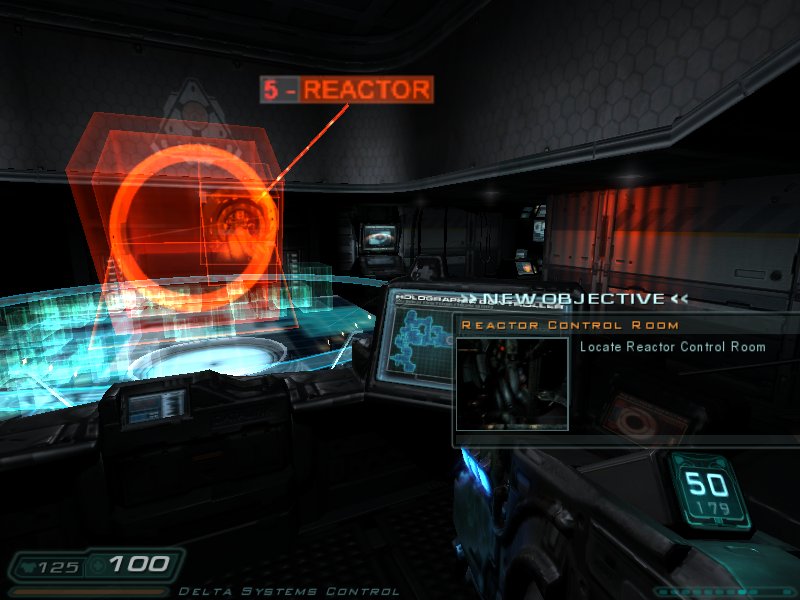
|[[Image:doom3.jpg|thumb|Doom 3|150px]] | |||
|[[Image:quake4_2.jpg|thumb|Quake 4|150px]] | |||
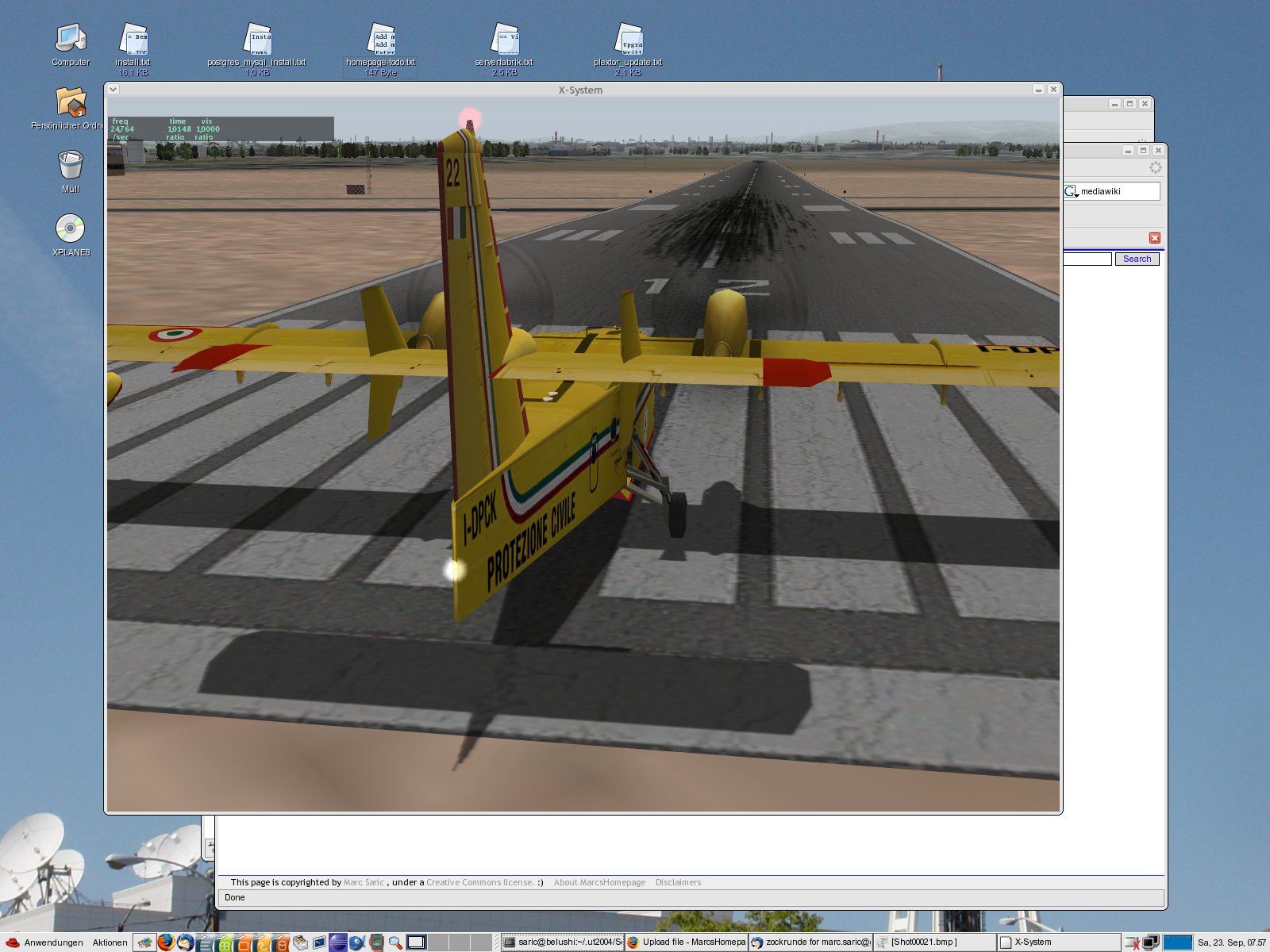
|[[Image:x-plane.jpg|thumb|X-Plane 11|270px]] | |||
|} | |||
There are some decent games out there, which run natively on Linux (see above :-) ). | |||
With [http://www.geek.com/games/gabe-newell-steam-on-linux-a-response-to-the-coming-windows-8-catastrophe-1505337/ Steam-support] the number of games natively supported on Linux has been quite OK recently, although some already [https://www.reddit.com/r/linux/comments/17x0sh/john_carmack_asks_why_wine_isnt_good_enough/c89sfto wrote it of] and -of course- many block buster releases are Windows (or console) only. | |||
Running Windows games on Linux is mostly not fun, so I ditched that and run Windows games on Windows nowadays. | |||
= Android = | |||
And of course, there is that [[Sony Xperia Z5|other Linux computer]], I carry around. | |||
Revision as of 18:35, 18 December 2024
My Linux-experience
I have been using Linux (mostly in a dual-boot environment) since around 1996 or 1997 (Cheapbytes CD from Lehmanns bookstore with Debian 1.24 on a Pentium 133 MHz).
The most recent version of this page has been created with Linux tools alone and is hosted on a Linux box. Some images and older sediments of my site may incorporate Windows-generated images, but nowadays I have switched to Open Source tools to create everything I want to show here.
I am developing Cloud-based software fro a living since my Postdoc days on Linux- and previously Apple-machines, using Python, Terraform, Ansible, Docker, Javascript, Mozilla, PHP, Perl,, PostgreSQL, MySQL and various IDEs.
Beside that I was heavily depending on a stable machine to do my Ph.D., which was one of the two main reasons for switching from Windows NT to SuSE-Linux and finally Fedora and Ubuntu.
Pictures
Because people always want to see screenshots here are some of my home-desktop. They are slightly recursive, the first one shows an editing-session of this HTML-page, running SuSE 7.3/KDE 2.2 with an IceWM-theme, the second one shows the upload of the homepage, running SuSE 8.1/KDE 3.0 with the Quartz theme and some nice tools. The third one is SuSE 9.1 KDE 3.2.1 and Gimp 2.0 the next is from the Fedora Core 3 installation on my AMD Athlon64 running Gnome 2.8 followed by a few shots from the FC6 install with 3D effects enabled. Next Fedora 10 Linux and Fedora 13 Linux and CentOS 6 and currently Ubuntu 14.04 LTS Desktop. Apparently I like blue backgrounds...
Raspberry Pi
I got a Raspberry Pi 4 with 2 GB RAM as a gift. Of course, I have to find some useful project for this little machine.
Read my Raspberry Pi project page for this.
Installing Linux on Laptops
Recently, I bought a used Lenovo Thinkpad x230.
Read the review.
During my work for Exelixis, I had an IBM R32 laptop running SuSE 8.1 and during my stay back at the university, I had a Toshiba M30-344[pdf-file], which I used with SuSE 9.0 and 9.1.
Short note: Both pages will not get updates, as I don't own those laptops any more.
Blue pill or red pill...
Gentoo Linux on Small Systems
Installing one of the more recent distros on an old system (aka my 300 MHz PII) is not really fun.
Except for the fact, that a small hard disk is quickly filled up by a load of gunk you might not need at all (but which is installed anyway, if you don't select rpm-packages by hand), those distros tend to be painfully slow (the current version of Quanta IS dead-slow even on my AMD Duron 700 (yess, CPUs have arrived ad 6.0 GHz meanwhile, I know)), and take eons to load the desktop (why? I was running Linux on my 486 ages ago).
There seems to be no real hope for a fast-and-small version of SuSE, therefore I had to look for something else.
Enter Gentoo-Linux: (I have to admit, that almost everything which follows, has been stolen from an email I got from a friend, who setup all this on his machine ;-)).
Anyway, if you are interested in my installation-notes for a small-footprint Gentoo-system, read on.
Fedora
Several years ago I switched from my long-time favorite Linux-distribution to Fedora Core 3.
Installation notes for Fedora Core 3 Linux
Permian period. My first stab at Fedora Linux after I switched from SuSE. Quite some manual effort here compared to more recent distros.
Installation notes for Fedora Core 6 Linux
Jurrassic age. One of the more unstable Fedora releases.
Installation notes for Fedora 8 Linux
Stone age. A better one (compared to the previous install).
Installation notes for Fedora 10 Linux
Very outdated. Some major havoc with Sun Java. Apart from that, the install becomes more and more boring mainstream
Installation notes for Fedora 13 Linux
Outdated. New install, new glitches, among them one, which cropped up in the current Ubuntu install.
Installation notes for Fedora 14 Linux
The last Fedora. I switched to CentOS afterwards.
CentOS
Installation notes for CentOS 4.3
Some time ago I had to setup a 64-bit 4-way server and decided to give CentOS a shot. Having ordered a floppy-drive ("who really needs that nowadays") immediately paid off, because the RAID-Controller was not recognized and a newer kernel-module had to be installed from a boot-floppy (not a problem, unless you can't use one).
Installation notes for CentOS 6.2
After everyone decided, that the new GUI for Linux should look like a Smartphone, I moved from Fedora to CentOS 6 (which also has a longer life-time and does not force you to install a new OS every 1,5 years or so).
Ubuntu Linux
Installing Ubuntu 7.10
Read my review on installing Ubuntu 7.10 Gutsy Gibbon. I did this recently for one of my older machines, because the old Ubuntu-version was no longer supported.
Installing Ubuntu 12.04 LTS 64 bit for XBMC
I have built a XBMC based Home Theater PC (HTPC) using Ubuntu 12.04 LTS 64 bit on an Intel platform.
Read my XBMC HTPC installation notes.
Installing Ubuntu 14.04 LTS 64 bit
My last PC died from Capacitor plague after more than 6 years, so I had to buy a new one. CentOS 7 was not quite ready, so I choose the new Ubuntu LTS. Read my Ubuntu 14.04 LTS Desktop installation notes.
Upgrading to Ubuntu 16.04 LTS 64 bit
As the update client kept nagging me, that a new version is out, I followed that advice and updated the existing system to Ubuntu 16.04 LTS. Read my Ubuntu 16.04 LTS Desktop upgrade notes.
Upgrading to Ubuntu 18.04 LTS 64 bit
I did an upgrade to Ubuntu 18.04 LTS. Reading my Ubuntu 18.04 LTS Desktop page is worthless, as it is empty (nothing to fiddle around with there).
Upgrading to Ubuntu 20.04 LTS 64 bit
I did an upgrade to Ubuntu 20.04 LTS. There is nothing to read there Ubuntu 20.04 LTS Desktop.
Gaming
There are some decent games out there, which run natively on Linux (see above :-) ).
With Steam-support the number of games natively supported on Linux has been quite OK recently, although some already wrote it of and -of course- many block buster releases are Windows (or console) only.
Running Windows games on Linux is mostly not fun, so I ditched that and run Windows games on Windows nowadays.
Android
And of course, there is that other Linux computer, I carry around.